DNA, The Fundamental Building Block of Our Genetic Makeup, has been intense nanotechnology Research Field. DNA nanotechnology is based on the idea that dna can be used as a programmable structural material.
Nucleotide Interaction is Highly specific following the Rule of Watson-Crick Base Pairing (IE, A-T and GC Paring), Which Allows rational placement and Assembly of DNA Strands Relative to each Other. DNA Strands also contain Chemically TURABLE Groups, which make it easy to functionalize dna with molecules and nanomaterials. AS A Result, DNA Nanostructures CAN ECODE BOTH STRUCTURAL AND CHEMICAL/Functional Information. More importantly, for the purposes of surface patterning, the cost of dna nanostructure is not a major concern.
Nanotechnology researchers use it to create artificial rationally designed nanostructures for various applications in biology, chemistry, and physics. DNA COULD Be Particularly USEFUL for the Design of Electric Circuits and DNA-Based Computing BECAUS IT SELF-AMOURS, IT SELF-REPLICATES AND IT CAN Adopt Various States and Conformation.
Compared to the Long-Term Storage Method that Uses Pizza-Sized Reels of Magnetic Tape, DNA Storage is potentialy less exisseed, Far More Physically Compact, More Energy Efficient, and Longer Lasting-DNA Survives for Hundreds of Years and Doesn't Require Maintenance. Files Stored in Dna Also can be very easyy copied for Negligible Cost.
DNA's Storage Density Is Staggering
Conside this: Humanity Will Generate An Estimated 33 Zettabytes of Data by 2025 - That's 3.3 Followed by 22 Zeroes. All that information WOULD FIT INTO A PING PONG BLL, with room to spare. The Library of Congress has about 74 Terabbytes, or 74 million million bytes, of Information - 6000 Such Libraries Would Fit in a Dna Archive the size of a poppy seed. Facebook's 300 PETABYTES (300,000 terabbytes) COULD BE STORED IN HALF A POPPY SEED.
At present, an Ever-over-rounding Number of Research groups are programmable exploitation self-assessly properies of nucleic acids in creating rationally designed nanoshapes and nanomachines for many different.
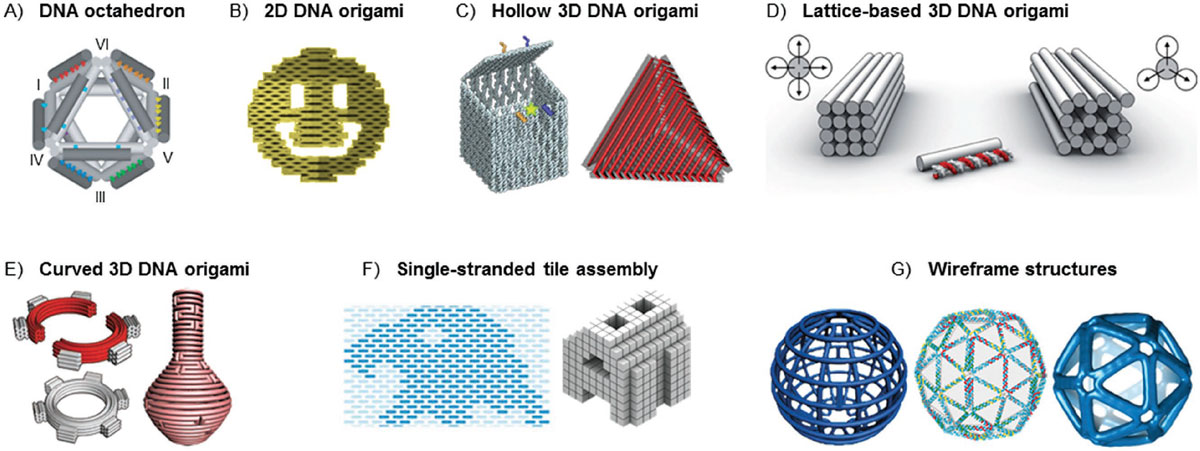
In A Progress Report in Advanced Materials, Scientists from Aalto University in Finland Focus on the Different Design Paradigms (DNA Origami and the Related Techniques Are Particularly Emphasized), Selected High-Quality Shapes, and the Software that Enable User-Friendly Design and Manufacturing of DNA Nanoobjects.
One of the Most Important Developments in Structural DNA Nanotechnology has been the use of A Scaffold Strand and Hundreds of Short Staples for the Assembly of Three-Dimensal (3D) DNA Origami Objects. These objects are similar in size to virus capsids and have gained a lot of interest as nanocontainers for drug deliver. Since they can be programed to carry out specific tasks (like Transporting Molecules and Rereshing Them at A Target Site), These Assemblies Act Like A Robotic System - Nanorobots.
The Review Paper Features An Extensive Discussion On the Origin of Dna Origami, Its Evolution from 2d to 3d, and the Trend Towards Building Larger Structures Beyond the Nanoscale.
The First Design Software for DNA Nanostructures WAS WRITEN BY NED SEEMAN in 1985 (Journal of Molecular Graphics, "Interactive Design and Manipulation of Macro-Molecular Architecture Nucleic Acid Junctions") and Succeeded by A Better Algorithm He Developed in 1990 (Journal of Biomolecular Structure and Dynamics, " Novo Design of Sequences for Nucleic Acid Structural Engineering ”).
In 2006, Ned Seeman and Colleagues Developed Gideon, Which Was Later on Partially replacement with other software and techniques - However, Gideon is Still Used in Many Labs. After the Dawn of DNA Design Software, the Researchers Quickly Adopted New and Revolutionary Dna Origami Method, and subsequently, this effort yaielded first origami-oriented software ssse and soon after even More Advanced Tools.
Cadnano by Douglas et al. and Cando by Castro et al. and Kim et al. Are widely used Together for Designing 2D and 3D DNA Origami Nanostructures and Computationally Predicting the Actual Shapes of the Designed Structures in Aqueous Solution. The core purpose of cadnano is to simplify and speed up the design process of dna nanostructures. Cando is a tool for predicting appearance, mechanical fluctuations, and flexibility, as well as twists and bends of the particular shape in the solution. The Software was developed by Hendrik Dietz's (Technische University München) and Mark Bathe's Groups (MIT) and it is Currently Sentained by the Laboratory for Computational Biology and Biophysics at Mit.
The Authors also discussion of tiamat, vhelix and daedalus programs.
With look to practical applications, the researchers bond that, despite the rapid progress in designing versatile structures and in developmental manufacturing methods reported in their paper, many implementations of dna nanotechnology are still quite impite.
However, there Already Exists implementations for helping researchers to map the Atomic Structure of Proteins, Computing in Living Cells, and Soon, Prospectively, Tracking and Curing Diseases with DNA Might Become Structures possible.
In the review's final section, the Authors Summarize the latest Advances in the Design Optimization and List Some Recent and notable Examples How Dna Nanotechnology can be harnessed in a number of subfields of Materials Science, Biotechnology, and Medicine.
In Particular, they remark on the Combination of Top-Down With Bottom-Up Nanofabrication to Fabricate Biosensors, Nanopores, Optics, Plasmonics, and Molecular Electronics; Molecular Computing, Signaling and Robotics; and finally nanobiomedicine and characterization of pivotal biomolecules.
